UK to create world-first 'early warning system' for pandemics (Concerns, perspectives & alternative approach)
This article will share my concerns and perspectives in relation to the UK's government proposal for the creation of a 'world-first early warning system for pandemics' [1].
In their document, the following claims are made:
It uses samples from patients with severe respiratory infections and rapid genetic testing to match those patients with the right treatments within 6 hours.
To address the current time lag between new pathogens emerging in the UK and action being taken to both treat affected patients and to prevent their spread.
In my view, given the past experience with the covid-19 event, this initiative might introduce the risk of misrepresenting sequencing data, offering novel treatments and/or countermeasures that will only harm public health. We need to understand that viruses and bacteria are part of the ecosystem, not biological hazards, and err on the side of caution with regards to novel diagnostics and widespread testing, as well as the introduction of novel treatments and changes in protocols during perceived emergency situations.
In relation to metagenomic sequencing via the use of nanopore technology or other devices, pathogen identification and establishing causation are not straightforward steps. Many signals can be identified via sequencing and misinterpreted as the primary etiological factor, leading to overdiagnosis and/or misdiagnosis, regardless of efficiency compared to traditional culture techniques.'
Alternative approaches for the use of metagenomic sequencing in health care settings (considerations for a meaningful definition of 'outbreak of disease'):
Outbreaks cannot become an international issue unless several regions share a series of conditions that make them susceptible to proliferation of disease. Microbes constantly circulate throughout the ecosystem, therefore disease states do not move via transmission chains, in the traditional sense, but rather via gradients of non-equilibrium states, were the same set of pathogens might transition from a disease phase to a non-disease phase based on several different interrelated signaling pathways within the holobiont, at the individual level and at the community level [2][3][4]. The primary use for metagenomic analysis should be to broadly characterize the background levels of such patterns so we can actually infer where a real outbreak is actually taking place. Considering a positive for the presence of a given genomic footprint (be it a newly identified one or not), cannot be applied for the detection of disease without the broader ecological context, unless we want to risk misinterpreting causality as positivity and produce harm via misdiagnosis, overtesting, overdiagnosis, and inadequate treatment protocols. The system they are proposing via the UK-nanopore collaboration, in its current formulation, doesn't seem to guarantee that appropriate mechanisms will be followed in order to properly characterize the clinical relevance of the measured signals. If they find high prevalence of a given pathogen in a severe case of disease, they will instruct to follow a contention mechanism and all positive detections of that given molecular print within the population, regardless of clinical significance, will be considered a threat, causing severe public health disruptions and generalized harm, while not only not preventing any outbreaks but creating imaginary ones. Their project is potentially dangerous and hazardous for public health. When such overreaction mechanisms are shared internationally, a Public Health Emergency of International Concern (PHEIC) could be declared for no reason whatsoever and the detection of the given molecular pattern will be guaranteed, since most microbes are already present at different levels of prevalence within different ecological niches and population cohorts in different phases of dynamic equilibrium. However a highly complex network of interrelated biological processes, of multi-causal origin, is the determinant of disease and not the presence or absence of genomic footprints on their own.
Hence, the best approach with regards to the possibilities offered by this new technology, would be to properly characterize the background signal, so we can compare it with outbreaks (if they occur), in order to see what elements are related to the imbalance in the ecosystem and what constitutes normal levels. Pathogens do not cause disease states by themselves, it is a highly interdependent dynamical system. If instead of characterizing the background and the different phases, from healthy to unhealthy, within individuals and at the population level, we solely focus on detection; normal levels, after the outbreak has already ended, would still be considered a risk, leading to misdiagnosis and harm. Characterizing the background would also help in understanding where the outbreak is actually occurring, which might be a very restricted area with a very specific set of conditions, rather than every region with molecular positivity. We would also be able to see how the same elements relate to each other and how different patterns, or different proportions of several microbes, are associated with health or disease, intermittently, and what the true etiological factors are. The etiological factors would always be ecosystemic in nature and multi-causal. It might be related to humidity, temperature, certain types of food, certain behaviors, low hygiene, economic income, poverty levels, mold, contaminants, air quality... never the presence or absence of a given genomic pattern.
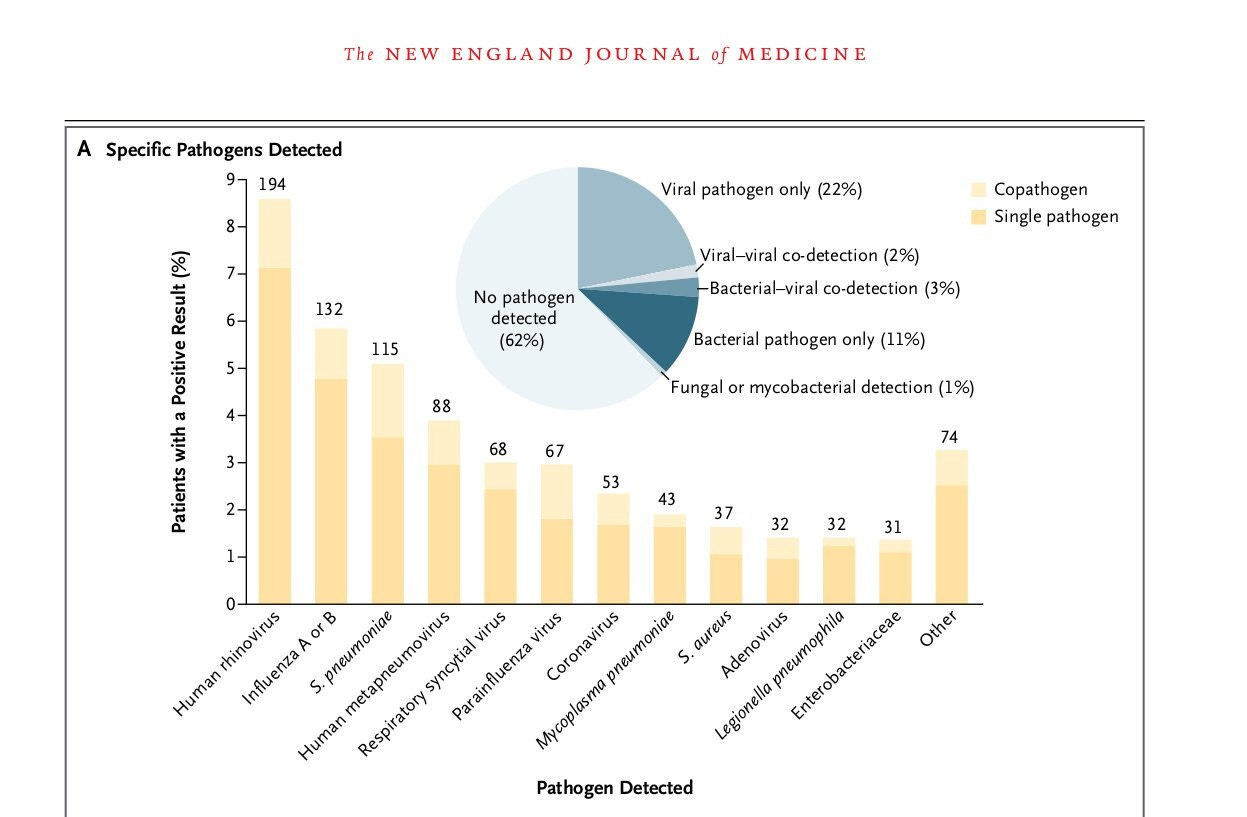
The severe consequences of widespread testing of any given molecular signal without considering its true clinical significance are highlighted in this hypothetical scenario below. Note: The CDC’s EPIC studies and the highlighted results, in screenshot format, are referenced in [5][6].
Concerns regarding potential ulterior motives:
If public health were the main concern of everyone involved in such projects, we might be able to improve such systems in order to avoid false alarms. However there are many interests and potential ulterior motives, such as political gain via the abuse of emergency powers, global governance via fear and control, economic interest within the pharmaceutical industry, the inappropriate promotion of experimental gene-based nanotechnologies for the purpose of changing public perception, habits, needs and biases... The dangers are severely concerning, and thus, the precautionary principle should rule against any such, so-called, 'early warning systems for pandemics'. The very use of the word 'pandemic', instead of outbreak or epidemic, makes it even more concerning, as if there is already a whole industry building around the concept of pandemics; and that is extremely dangerous.
There are also serious implications regarding the misuse of emergency powers and PHEIC declarations.:
References:
[1] UK to create world-first 'early warning system' for pandemics https://www.gov.uk/government/news/uk-to-create-world-first-early-warning-system-for-pandemics?utm_campaign=&utm_source=threads,twitter&utm_medium=social&utm_content=1737019487
[2] The virome in mammalian physiology and disease https://pubmed.ncbi.nlm.nih.gov/24679532/
[3] The Intra-Dependence of Viruses and the Holobiont https://pubmed.ncbi.nlm.nih.gov/29170664/
[4] Humans as holobionts: implications for prevention and therapy https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0466-8
[5] Community-acquired pneumonia requiring hospitalization among U.S. children https://pubmed.ncbi.nlm.nih.gov/25714161/
[6] Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults https://pubmed.ncbi.nlm.nih.gov/26172429/















Some interesting references there.
What I don't get is this: given the content of those references, in what way could metagenomic sequencing ever be a good thing in light of our lack of understanding of all the elements of this jigsaw.
Surely it is inevitably going to be harmful.
So I don't understand why you say:
"Hence, the best approach with regards to the possibilities offered by this new technology, would be to properly characterize the background signal, so we can compare it with outbreaks (if they occur), in order to see what elements are related to the imbalance in the ecosystem and what constitutes normal levels. "